1 Scientific documents
In their daily work, students, academics and scientific specialists produce a large amount of documentation of all kinds: laboratory notes, lectures, memos, technical reports and, above all, scientific articles to publish their discoveries and advances in an area of knowledge. Typically, creating this type of scientific document involves many tasks involving different tools and possible points of failure.
Figure 1.1 shows a schematic overview of a classic workflow for creating scientific documents. The main element is often a word processor master file (like Word or OpenOffice/LibreOffice), a web page, or a LaTeX file (if we are creating a PDF document) that holds all contents.
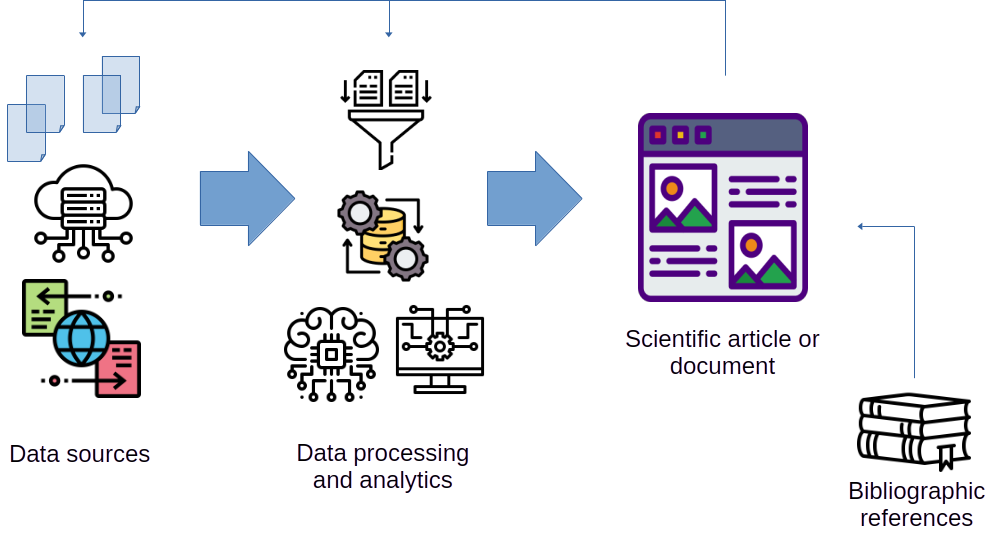
This master file is filled with content from a variety of sources, such as:
- figures and diagrams generated manually or through software code (such as data visualization charts);
- tables and summaries describing data sets and results;
- results and evaluation of the performance of models or algorithms; statistical or machine learning;
- mathematical formulas and equations;
- data tables and other useful information;
- bibliographic references (usually generated with the help of some bibliographic information management program).
Many of these elements force users to run external tools and programs, procedures, and other tasks over and over again and then incorporate the new results into the master file. We must admit that this process, which is mostly manual, is tedious and very prone to errors or oversights. “Wait! I forgot to update Figure 1.” “Are you sure these are the latest evaluation results for model M?” “Have you checked if we have uploaded the latest version of the data file D?” These are common questions that arise in the day-to-day work of scientific teams.
However, it would be great if it were unnecessary to carry out all this manual and sometimes very frustrating process manually. Do we have any alternative to avoid it? Yes, we do. The answer to our needs is provided by a compelling concept: literate programming.
1.1 Literate programming
In 1984, professor Donald E. Knuth (1984) coined the concept of literary programming. Yes, that happened more than 40 years ago. This concept states that it should be possible to integrate formatted text and results of the execution of software code to compose said document dynamically in a single scientific document. So, why has it taken us so long to implement this idea? Knuth’s vision, although significantly ahead of its time, was correct, but the technology of the time did not allow it to become real.
However, we have all the essential elements to make it real today. Quarto is a software tool that lets us automate and manage the entire process of creating literary programming documents quickly and reliably.
1.2 Reproducible research
For many decades, the scientific method has relied upon publishing research papers that describe the results of data analysis and experiments. In all cases, it is essential to be able to trust the conditions, the data collected, the method of analysis and execution of the experiments, along with the various kinds of tools, including software, that the authors of the publication used to carry it out.
However, the numerous advances in recent years in the tools and methods of analysis make it much easier to check the results of these analyses. We might assume that this makes the work of scientists much easier, but in reality, the opposite is true. Let us look at some examples:
Oncology (Begley & Ellis, 2012): The Biotechnology Department of Amgen (Thousand Oaks, CA, USA) was able to confirm only 6 of a total of 53 emblematic research articles published in this area. Bayer HealthCare (Germany) was able to validate only 25% of the studies analyzed.
Psychology (Wicherts et al., 2006): 73% of the authors of 249 articles published by the APA did not respond within 6 months to the questions and requests formulated about the data they used in their research.
Economics and Finance (Burman et al., 2010): A comparison of different software packages applied in the execution of various financial and statistical model analyses shows that each of these packages produces very different results using the same statistical techniques directly applied to identical data as those used in the original publication.
In fact, articles have even appeared suggesting that many of the results published in areas such as Medicine may not be entirely reliable (Ioannidis, 2005). As a result of all these recent findings, a great controversy has appeared throughout the scientific and research community, accompanied by a deep crisis of confidence.
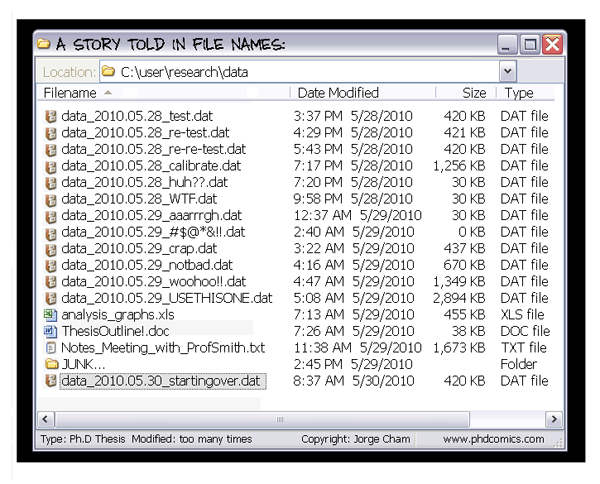
Nevertheless, as a well-known comic strip about the academic world and research (see Figure 1.2) very well describes, developing scientific publications is based primarily on the continuous review of methods and results (starting with the students themselves and their supervisors).
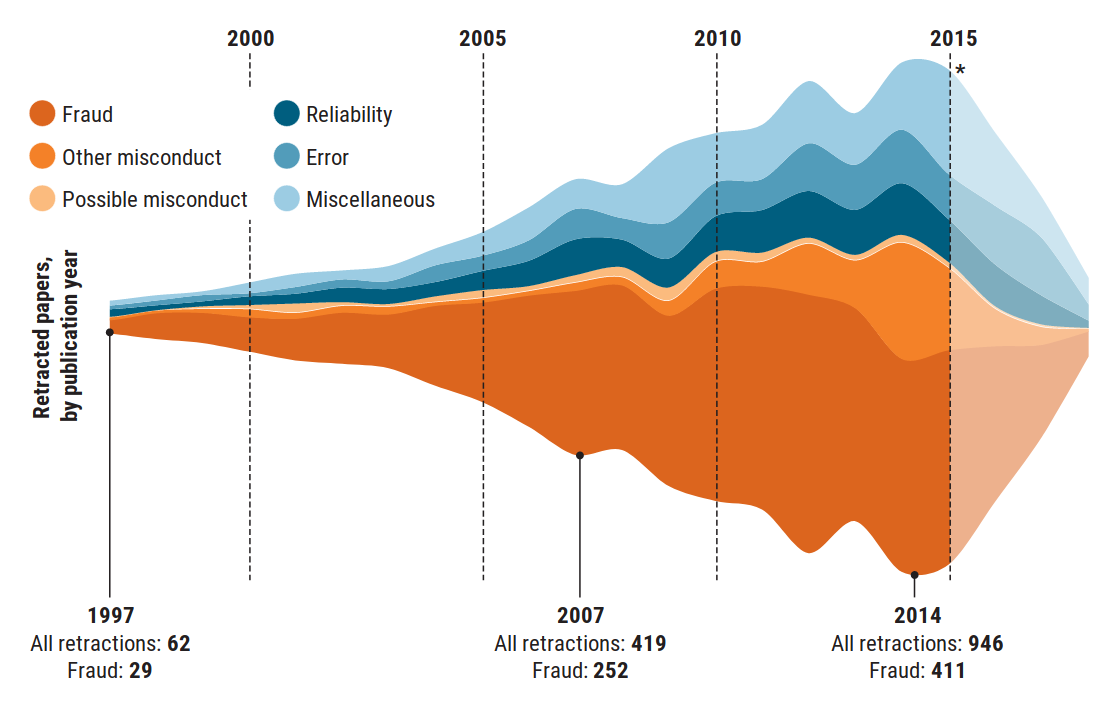
The Figure 1.3 shows a graph published in the prestigious journal Science Magazine (Brainard et al., 2018), representing the evolution of the number of research articles retracted or withdrawn for various reasons between 1997 and 2014. In this graph, we can see how the improvement of tools and the greater availability of resources allow for the analysis and review of a greater volume of publications and analyses, which allows for the detection of a more significant number of problematic cases.
1.2.1 Reproducibility and replicability
There is often talk of reproducing and replicating a data analysis or a scientific experiment (Leek & Peng, 2015). However, many shreds of evidence show incompatible definitions of these two and other related terms (Barba, 2018). Be very careful because depending on the scientific community or the field of knowledge we find ourselves in, the meaning of these two terms may even be opposite to their accepted definition in other areas 1. Here, we will stick to the definition accepted in a large number of areas, including statistics or scientific computing (see Barba, 2018, p. 33):
Reproducibility is the ability to recompute the results of an analysis with the same data used in the original analysis and know the sequence details (workflow or pipeline) of operations that make up said analysis. Certain premises must be able to be guaranteed:
If we use the same tools (e.g. R, a specific list of packages, identical versions of all packages and dependencies), as well as the same code (R scripts) on the same data, the results and conclusions must be consistent with those of the original analysis.
The authors of the original analysis must provide all the elements (data, code and procedure used) to allow the analysis to be reproducible (Barba, 2018).
Replicability is the ability to perform an experiment or analysis independent of the original that addresses the same objective but on a set of data different from that used in the initial study. If the results are not consistent, more replications will be necessary to harmonize the results and conclusions through appropriate techniques, such as meta-analysis.
1.2.2 Replication levels
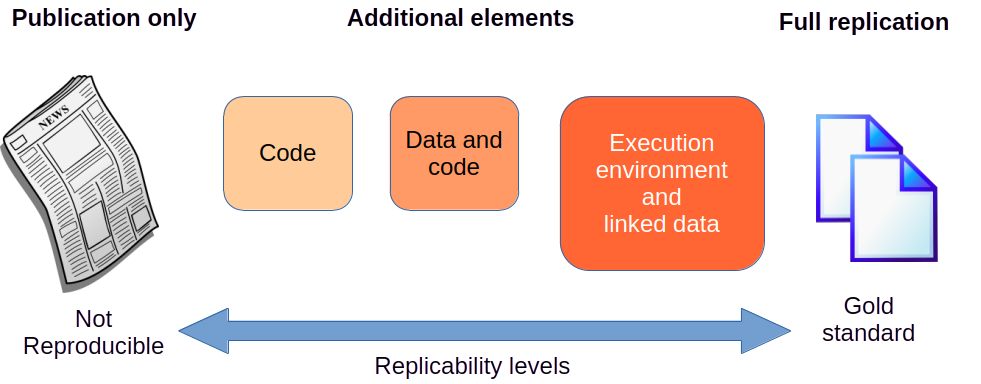
Depending on the elements published by the authors of the original study and the details provided regarding the process to carry out the study, its steps, and involved tools, we have different levels of replicability or reproducibility, represented in the Figure 1.4.
Not reproducible: No data, code or specific description of the implementation of the study or analysis is available. Many scientific publications no longer accept publishing articles under these conditions.
Code or Data: A good number of publishers request that the data sets used in the analysis or study of the publication be accessible through a URL, either because they are available in a public repository or because the authors of the article have published it. Likewise, many publications require that the software code to carry out the analysis is also publicly accessible in an open source repository or a freely accessible version control service project.
Code and data: Ideally, both the code and the data should be publicly accessible for anyone who wants to examine them or use them to reproduce the results (validation) or replicate the analysis with other data or other cases.
Runtime environment and linked data: A further step to facilitate the reproducibility of studies consists of publishing code and metadata files with more precise information about the programming language, the software packages used and any other dependencies necessary to carry out the same study or analysis. Another variant to facilitate reproducibility is encapsulating the code and dependencies in a preconfigured virtual container to download and execute it directly.
Gold standard: The most advanced level would consist of documenting all the procedures performed during the study or analysis, including coding tasks of obtaining, cleaning and preparing the data, and producing graphics to visualize the results or any other results derived from the study.
1.2.3 Replicability tools
Specific technologies and tools that have become more sophisticated and refined in recent years are making replicating data processing and analysis easier.
- Version Control Systems for software code (SCV): Tools such as Git, Mercurial and web services such as GitHub or GitLab have popularized the creation and publication of projects, enabling software code management and controlling the changes and the released versions. Web services also integrate a good number of tools to support different facets of the software development process, such as the generation of documentation, manuals and examples, error reports and requests for improvements, continuous integration and continuous deployment (CI/CD), systematic testing of the generated code, etc. If you have not yet considered how using a tool for code version control can benefit you, take a look at Figure 1.5, where you will relive a situation that is unfortunately very common among researchers and scientists who develop software solutions.
Software virtualization and containers: In a technological environment dominated by the contracting and deployment of computing infrastructure and services in cloud computing architectures, packaging and virtualization tools for software applications and services that can be installed and deployed in a short time have revolutionized the way software products are published and managed, including data processing and analysis products.
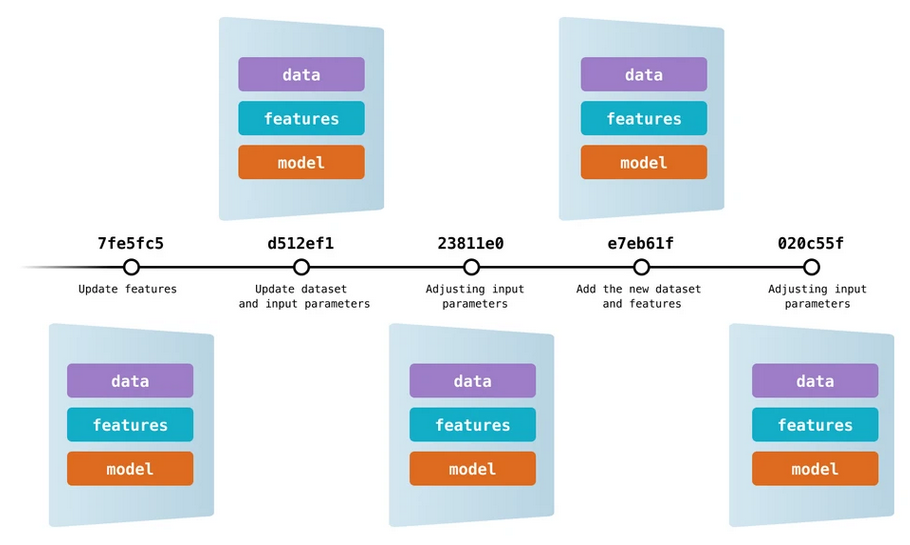
Data version control: Akin to SCV for source code, new software tools bring the same principles to data files. This way, we can control different data file versions, modifications, etc. Data Version Control (DVC) allows versioning of data and models. As a result, we can always know which version of the data and which list of features have been included in each model considered during the analysis, keeping the descriptive information about these three essential components that must always be cohesive.
Model and experiment management: Another type of project management tool for machine learning projects allows the organization, monitoring, comparison, and selection of the experiments and models we have carried out. One of the most recent notable examples is ML Flow, which provides support for model tuning, evaluation, and optimization, deployment of models in production environments, creation of a registry of pre-trained models, etc. Of course, combining this type of tool with others, such as DVC, can create a comprehensive management environment for our projects.
Creation and management of data processing pipelines: The last essential element in any data processing and analysis project that must take care of scalability is a tool for creating and managing data processing and analysis flows or pipelines. The set of all the pipelines in our project makes up the general workflow of the project. These tools are known as data or workflow orchestrators. In this category, we have compelling and feature-packed tools such as Apache Airflow or Prefect, and simpler, more straightforward ones such as Luigi.
Of course, the R community has not remained oblivious to these new trends, in particular the R OpenSci initiative, within which we find many packages (published in the official CRAN repository) that cover various aspects of scientific work, including pipelines and workflows management through the targets package.
1.3 Quarto for scientific publications
Now that we know the fundamental concept on which Quarto works and its application to achieve a higher level of reproducibility and transparency in our scientific process, we wil explain in more detail the process that Quarto follows to compose a document. The Figure 1.7 presents a diagram with the document creation process and the elements and tools that come into play to achieve it.
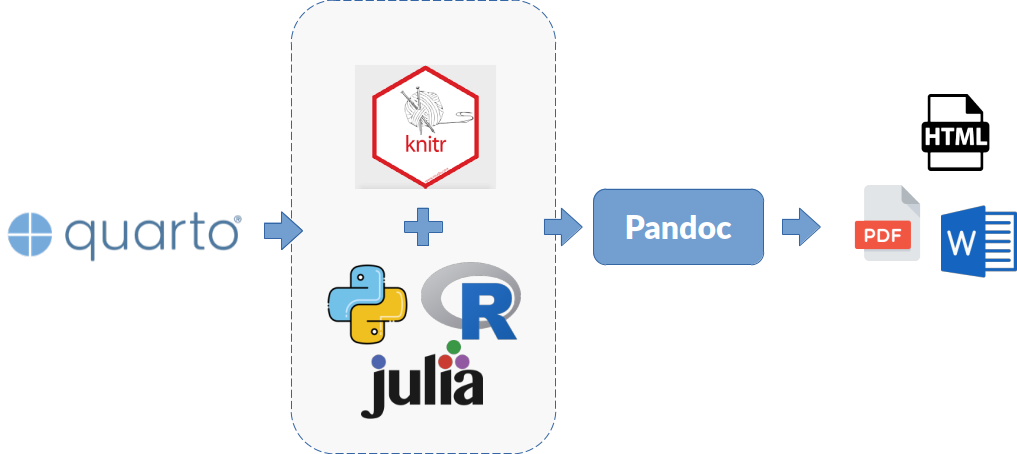
- Quarto: A software to create scientific documentation following the principles of literary programming.
- Knitr and programming language: The
knitrpackage is responsible for the connection with an interpreter of a programming language (R, Python, Julia) that can be executed in a REPL environment, in order to be able to execute software code fragments integrated into the document and generate content in Markdown format as a result. - Markdown (formatted content): a textual content markup language that allows easy formatting of the information in our documents created with Quarto.
- Pandoc (universal translator of document formats): This software receives content already formatted using the Markdown standard and converts it into the selected output type. Several options are available: HTML, PDF, Word, slides, websites, or interactive panels (dashboards).
1.4 Quartion installation
To install the latest version of Quarto software on your system, point load the URL https://quarto.org/docs/get-started/ in a web browser. Here, download and install the file corresponding to your operating system.
Currently, the latest version of Quarto available is 1.5.57.
By default, the output format of documents generated with Quarto is HTML. If we want to generate PDF documents, we must install a LaTeX distribution. For more information, see Section 3.3.
Among the most important examples of definitions that contradict those we give in this workshop are those adopted by the Federation of American Societies for Experimental Biology (FASEB) in immunology and microbiology, as well as those adopted by the Association for Computer Machinery (ACM) in computer science.↩︎